 Requirements of Project
Requirements of Project
Requirements
Instructions
Instructions
Marking Guide
Dataset
BRCA-50 is a Breast cancer dataset, including the expression levels of 50 important genes in Breast cancer.
- The dataset includes 1212 samples with
- 112 samples are of normal cases (class = N) and
- 1100 samples are of cancer patients (class = C).
Tasks
Use a causal structure learning algorithm to find the gene regulatory network, i.e. the network showing the interactions between genes, using the gene expression data. Explain how the algorithm works. (4)
Hints
Hints: Please exclude the class variable in building the network
EBF1is an important gene that is involved in many biological processes leading to cancer. Find the top 10 other genes that have strong causal effects onEBF1using a causal inference algorithm. (4)Hints
- Exclude the class variable in building the network
- If there are multiple possible causal effects between the cause and the effect, we can use the minimum of the absolute values (of the causal effects) as the final result
- The causal effects are normally ranked based on their absolute values.
Use a local causal structure learning algorithm to find genes in the Markov blanket of
ABCA9from data. Explain how the algorithm works. (4)Discretise the dataset to binary using the average expression of ALL genes as the threshold. The discretised dataset will be used in the following questions.
Use PC-simple algorithm (pcSelect) to find the parent and children set of the class variable. Explain how PC-simple works.
- Evaluate the accuracy of the Naïve Bayes classification on the dataset in the following cases:
- Use all features (genes) in the dataset
- Use only the features (genes) in the parent and children set of the class variable
- Compare the accuracy of the models in the two cases using 10-fold cross validation. (6)
- Evaluate the accuracy of the Naïve Bayes classification on the dataset in the following cases:
Given a Bayesian network as in the below figure
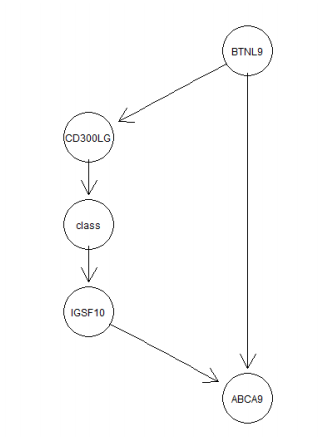
- a) Construct the conditional probability tables for the Bayesian network based on data. (3)
- b) Estimate the probability of the four genes in the network having high expression levels. (2)
- c) Estimate the probability of having cancer when the expression level of
CD300LGis high and the expression level ofBTNL9is low. (2) - d) Prove the result in c) mathematically. (2)
- e) Given we know the value of
CD300LG, is the “class” conditionally independent ofABCA9? And why? (3)
