 Assignment1
Assignment1
Requirements
Instructions
Instructions
Marking Guide
Part 1
Requirements
a) Describe a big data application that the company is using and use it to illustrate the characteristics of big data analytics. (4)
b) Suggest and describe one new big data application that would help the company improve their business performance. (4)
c) Explain why your suggested application is innovative and useful. Discuss the challenges of implementing the application that you proposed. (4)
1.1 Big Data Applications in Meituan
For a company that need to process big data, all they will come across the problems because of characters of big data. There are several ways to describe the characters of big data, such as the most famous V3 and V10. Next, will explore how the Meituan, which is a comprehensive internet company in China, to process big data for their business.
Here will use the V3 to show the characters that Meituan come across. Firstly, describe the huge volume of data, there are 42P+ volume data exist in company and around 16 thousand tables; for the variety, there are kinds of business data, log, and comments, etc. need to process; for the velocity, there are 150 thousand tasks on MapReduce and Spark per day, the log data with a peak value of one million per second. There are 2500+ nodes are deployed in 3 computer rooms to process all the tasks (2022).
It’s obviously that it impossible to process using a normal application. They established application based on Hadoop software to support all the service. The structure of their application like Image 1.1.1.
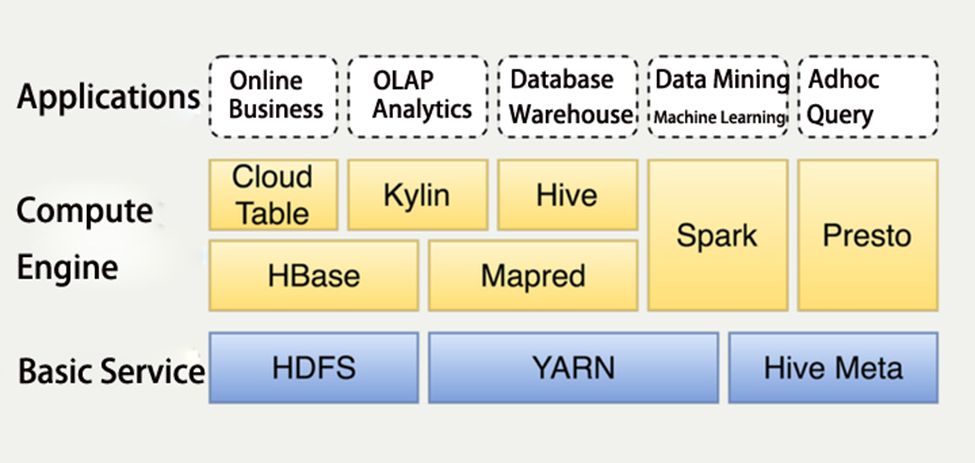
The applications layer is the business layer, they are all business related, the compute engine layer and basic service layer construct the big data platform, which are what we need to focus on. All the components on the two layers will be described below.
HDFS stands for Hadoop Distributed File System, which is a distributed file system (HDFS Architecture Guide, no date). Apache Hadoop YARN (Yet Another Resource Negotiator) is a novel resource orchestrator within the Hadoop framework, and a versatile resource governance infrastructure capable of furnishing cohesive resource allocation and scheduling functionalities for superjacent application strata (Apache Hadoop 3.3.6 – Apache Hadoop YARN, no date). The fullname of Hive Meta is Hive Metastore (HMS), which is the paramount depository, it houses the metadata pertaining to Hive tables and partitions within a relational database, affording diverse clients (comprising Hive, Impala, and Spark) the capability to retrieve said information through the utilization of the Metastore service API. (Apache Hive, no date). HBase is Apache HBase™ is the Hadoop database, a distributed, scalable, big data store (Apache HBase – Apache HBaseTM Home, no date). Mapred is a package, which describes how to read and write ORC files from Hadoop’s older org.apache.hadoop.mapred MapReduce APIs (Using in MapRed, no date). Cloud Table is a HBase interface make by Meituan (2022). Apache Kylin stands as an open source, distributed Analytical Data Warehouse; its conception was geared towards furnishing OLAP (Online Analytical Processing) proficiency within the milieu of extensive data landscapes. (Apache Kylin | Analytical Data Warehouse for Big Data, no date). Apache Hive is a distributed and resilient data warehousing system, which empowers extensive-scale analytics, querying, manipulation, and governance of petabytes of data stored across distributed repositories through the SQL. (Apache Hive, no date). Apache Spark is a multi-language engine for executing data engineering, data science, and machine learning on single-node machines or clusters (Apache SparkTM - Unified Engine for large-scale data analytics, no date). Presto is an open-source SQL query engine that's fast, reliable, and efficient at scale (Presto: Free, Open-Source SQL Query Engine for any Data, no date).
After having a glace of each component, next step will give some advice on how to optimize the big data platform.
1.2 Suggestions
The platform structure formed around 2012, ten years past, there are lots of components have made a great progress, and there are replacement components emerged. The structure of Meituan’s platform is well organized, but still have several components can be optimized. I will propose one suggestion on the structure, which is using K8 that is an open-source system for automating deployment, scaling, and management of containerized applications, to replace yarn for a more flexible capability (Production-Grade Container Orchestration n.d.).
1.3 The Advantages
This part will show the advantages for the suggestions proposed on chapter 1.2. For replacement using K8 for YARN, the first thing is to understand the details of the two components, and the limitations of YARN and advantage of K8.
For understanding the limitations of YARN, it’s necessary to know how YARN works. A YARN cluster consists of nodes, some of them are Master nodes, and the most Worker nodes. Two resource managers to manage resource at different levels. The ResourceManager handles resources at the cluster level, while NodeManager manages resources at the individual host level. They track vcores and memory at the cluster and localhost level. When an application runs on YARN, the two managers will evaluate the available resources, then assign each container to a host. In this way, the key work of YARN is to manage resources and schedule tasks on the cluster.
YARN exhibits limitations like version control, job isolation, and resource allocation. Running diverse workloads mandates separate clusters, escalating complexity and inefficiency. Especially for demanding tasks like real-time processing, YARN's lack of job isolation necessitates frequent cluster setup, causing costs and resource wastage (Kubernetes vs YARN for scheduling Apache Spark n.d.).
The next is to understand how K8 works and the benefits from using K8.
Kubernetes could use pod to manage different tasks as an isolated container, a pod is a group of containers, and all the tasks run in an isolated environment, no matter which task failed will not influence the whole cluster (Sensu | How Kubernetes works n.d.).
After comparation between the YARN and Kubernetes, several benefits will get after using Kubernetes. Such as, containerize applications and dependencies to prevent dependency issues; Kubernetes' Resource Quota and Namespaces enhance control over resource utilization; portable hybrid cloud compatibility achieved with swappable backends for Spark applications; Kubernetes Role and ClusterRole features enable precise permissions based on API groups; Tag container images for version control, aiding auditing and rollback of deployments; flourishing Kubernetes ecosystem offers robust open-source management add-ons like Prometheus, Fluentd, and Grafana.
All the benefits are the reasons for the suggestion.
1.4 Challenges
The Kubernetes is a great component for dealing with big data. There still have lots of challenges to overcome. The initial hurdle in adopting Kubernetes lies in the requisite expertise, which often lacking in data teams. Proficiency in Kubernetes, Helm, Docker, and networking basics is essential. Despite Kubernetes' prowess in scaling apps, addressing infrastructure scalability remains a task. Efficient cost management amid the need for adaptable infrastructure supporting dynamic applications is another significant challenge. Given the resource-intensive nature of big data tasks involving research, testing, modeling, and experimentation, costs can escalate if not vigilantly managed (Kubernetes vs YARN for scheduling Apache Spark n.d.).
Part2
For this part, the Weka will be selected as the tool to implement NaiveBayes Classifier, the original data need to convert into a proper format. The csv file format is selected, please download the data follow the link below.
Breast cancer wisconsin data (Have been converted into csv)
Numerical Variables
Part A: Model (3 marks)
Requirement
- Remove the ID attribute. Consider all attributes except the class attribute as numeric attributes.
Requirement
- Build a Naïve Bayes model and classify benign and malignant. Show the screenshot of the model when you train the model with all records.
Full Training Set
=== Classifier model (full training set) ===
Naive Bayes Classifier
Class
Attribute benign Malignant
(0.65) (0.35)
==================================================
Clump Thickness
mean 2.9563 7.195
std. dev. 1.6725 2.4238
weight sum 458 241
precision 1 1
Uniformity of Cell Size
mean 1.3253 6.5726
std. dev. 0.9067 2.7139
weight sum 458 241
precision 1 1
Uniformity of Cell Shape
mean 1.4432 6.5602
std. dev. 0.9967 2.5567
weight sum 458 241
precision 1 1
Marginal Adhesion
mean 1.3646 5.5477
std. dev. 0.9957 3.2038
weight sum 458 241
precision 1 1
Single Epithelial Cell Size
mean 2.1201 5.2988
std. dev. 0.9161 2.4465
weight sum 458 241
precision 1 1
Bare Nuclei
mean 1.3468 7.6276
std. dev. 1.1765 3.1102
weight sum 444 239
precision 1 1
Bland Chromatin
mean 2.1004 5.9793
std. dev. 1.0792 2.2691
weight sum 458 241
precision 1 1
Normal Nucleoli
mean 1.2904 5.8631
std. dev. 1.0577 3.3437
weight sum 458 241
precision 1 1
Mitoses
mean 1.1889 2.7401
std. dev. 0.4833 2.5138
weight sum 458 241
precision 1.125 1.125Part B: Explanation (3 marks)
Requirement
Use one record to explain how the model makes classification.
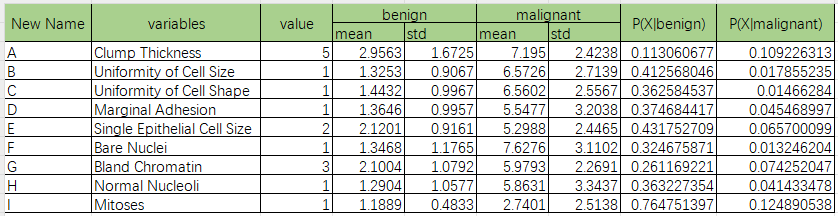
We select one record like the Image below as the test record, and show how the model works
According to Image above, we could know the value of each variable. For easily to use, the variables need to rename for short.
| New Name | Original Name | Value |
|---|---|---|
| A | Clump Thickness | 5 |
| B | Uniformity of Cell Size | 1 |
| C | Uniformity of Cell Shape | 1 |
| D | Marginal Adhesion | 1 |
| E | Single Epithelial Cell Size | 2 |
| F | Bare Nuclei | 1 |
| G | Bland Chromatin | 3 |
| H | Normal Nucleoli | 1 |
| I | Mitoses | 1 |
Now, We assign
Because of
Because of the conditional probability for continuous-valued features equals to
According to the model, we could know the mean and standard deviation like the picture below.
P(Class=benign) = 0.65
P(Class=malignant) = 0.35
Finally, we could calculate that
It's obviously that
So, the final result for this record is benign, it has been classifed correctlly.
Tips
If you want to calculate other record please use the Probability calculator.
Part C: 10 fold cross validation (3 marks)
Requirement
Show the accuracy of the model using 10-fold cross validation and the confusion matrix. Show and explain the meaning of the precision and recall for malignant.
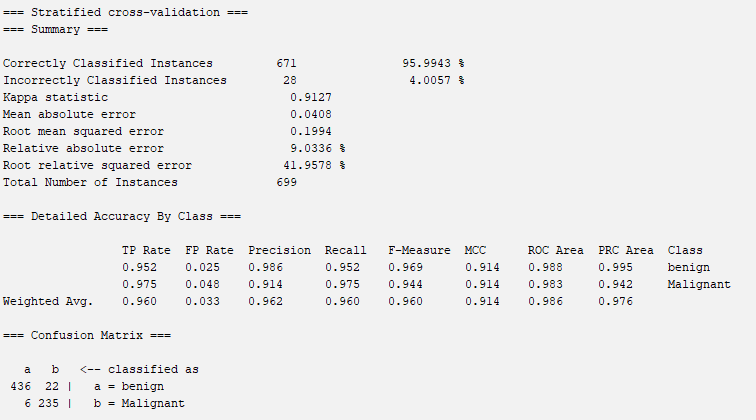
According to the results for cross validation, it is easy to get the confusion matrix and other indicators.
The accuracy of the model is about 95.9943%.
10 fold cross validation
=== Run information ===
Scheme: weka.classifiers.bayes.NaiveBayes
Relation: breast-cancer-wisconsin-weka.filters.unsupervised.attribute.Remove-R1
Instances: 699
Attributes: 10
Clump Thickness
Uniformity of Cell Size
Uniformity of Cell Shape
Marginal Adhesion
Single Epithelial Cell Size
Bare Nuclei
Bland Chromatin
Normal Nucleoli
Mitoses
Class
Test mode: 10-fold cross-validation
=== Classifier model (full training set) ===
Naive Bayes Classifier
Class
Attribute benign Malignant
(0.65) (0.35)
==================================================
Clump Thickness
mean 2.9563 7.195
std. dev. 1.6725 2.4238
weight sum 458 241
precision 1 1
Uniformity of Cell Size
mean 1.3253 6.5726
std. dev. 0.9067 2.7139
weight sum 458 241
precision 1 1
Uniformity of Cell Shape
mean 1.4432 6.5602
std. dev. 0.9967 2.5567
weight sum 458 241
precision 1 1
Marginal Adhesion
mean 1.3646 5.5477
std. dev. 0.9957 3.2038
weight sum 458 241
precision 1 1
Single Epithelial Cell Size
mean 2.1201 5.2988
std. dev. 0.9161 2.4465
weight sum 458 241
precision 1 1
Bare Nuclei
mean 1.3468 7.6276
std. dev. 1.1765 3.1102
weight sum 444 239
precision 1 1
Bland Chromatin
mean 2.1004 5.9793
std. dev. 1.0792 2.2691
weight sum 458 241
precision 1 1
Normal Nucleoli
mean 1.2904 5.8631
std. dev. 1.0577 3.3437
weight sum 458 241
precision 1 1
Mitoses
mean 1.1889 2.7401
std. dev. 0.4833 2.5138
weight sum 458 241
precision 1.125 1.125
Time taken to build model: 0 seconds
=== Stratified cross-validation ===
=== Summary ===
Correctly Classified Instances 671 95.9943 %
Incorrectly Classified Instances 28 4.0057 %
Kappa statistic 0.9127
Mean absolute error 0.0408
Root mean squared error 0.1994
Relative absolute error 9.0336 %
Root relative squared error 41.9578 %
Total Number of Instances 699
=== Detailed Accuracy By Class ===
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.952 0.025 0.986 0.952 0.969 0.914 0.988 0.995 benign
0.975 0.048 0.914 0.975 0.944 0.914 0.983 0.942 Malignant
Weighted Avg. 0.960 0.033 0.962 0.960 0.960 0.914 0.986 0.976
=== Confusion Matrix ===
a b <-- classified as
436 22 | a = benign
6 235 | b = MalignantCategorical Variables
Part D: Discretizetion (3 marks)
Requirement
Discretise the data set using three bins (equal-frequency).
The discretizetion should use equal-frequency technique to discretise the numerical data into 3 bins. The configuration file for discrete like the link. The configuration for equal frequency in Weka
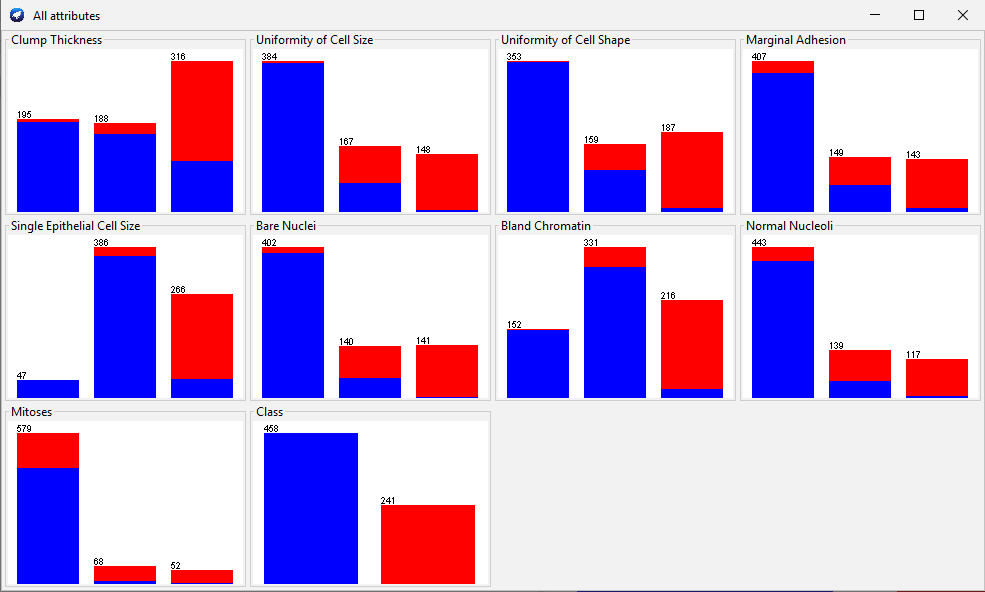
Part E: Model (3 Marks)
- Model and Evaluation
Requirement
Build a Naïve Bayes model and classify benign and malignant using the discretised dataset. Show the accuracy of the model using 5-fold cross validation and the confusion matrix.
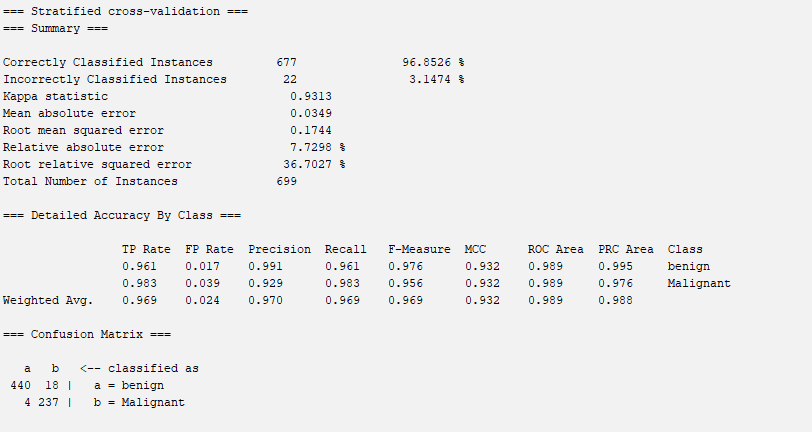
According to the result, it is easy to get the accuracy (96.8526%) and the confusion matrix.
Details
=== Run information ===
Scheme: weka.classifiers.bayes.NaiveBayes
Relation: breast-cancer-wisconsin-weka.filters.unsupervised.attribute.Remove-R1-weka.filters.unsupervised.attribute.Discretize-F-B3-M-1.0-Rfirst-last-precision6
Instances: 699
Attributes: 10
Clump Thickness
Uniformity of Cell Size
Uniformity of Cell Shape
Marginal Adhesion
Single Epithelial Cell Size
Bare Nuclei
Bland Chromatin
Normal Nucleoli
Mitoses
Class
Test mode: 5-fold cross-validation
=== Classifier model (full training set) ===
Naive Bayes Classifier
Class
Attribute benign Malignant
(0.65) (0.35)
==================================================
Clump Thickness
'(-inf-2.5]' 189.0 8.0
'(2.5-4.5]' 165.0 25.0
'(4.5-inf)' 107.0 211.0
[total] 461.0 244.0
Uniformity of Cell Size
'(-inf-1.5]' 381.0 5.0
'(1.5-5.5]' 74.0 95.0
'(5.5-inf)' 6.0 144.0
[total] 461.0 244.0
Uniformity of Cell Shape
'(-inf-1.5]' 352.0 3.0
'(1.5-4.5]' 99.0 62.0
'(4.5-inf)' 10.0 179.0
[total] 461.0 244.0
Marginal Adhesion
'(-inf-1.5]' 376.0 33.0
'(1.5-4.5]' 74.0 77.0
'(4.5-inf)' 11.0 134.0
[total] 461.0 244.0
Single Epithelial Cell Size
'(-inf-1.5]' 47.0 2.0
'(1.5-2.5]' 364.0 24.0
'(2.5-inf)' 50.0 218.0
[total] 461.0 244.0
Bare Nuclei
'(-inf-1.5]' 388.0 16.0
'(1.5-8.5]' 55.0 87.0
'(8.5-inf)' 4.0 139.0
[total] 447.0 242.0
Bland Chromatin
'(-inf-1.5]' 151.0 3.0
'(1.5-3.5]' 289.0 44.0
'(3.5-inf)' 21.0 197.0
[total] 461.0 244.0
Normal Nucleoli
'(-inf-1.5]' 403.0 42.0
'(1.5-6.5]' 50.0 91.0
'(6.5-inf)' 8.0 111.0
[total] 461.0 244.0
Mitoses
'(-inf-1.5]' 446.0 135.0
'(1.5-3.5]' 11.0 59.0
'(3.5-inf)' 4.0 50.0
[total] 461.0 244.0
Time taken to build model: 0 seconds
=== Stratified cross-validation ===
=== Summary ===
Correctly Classified Instances 677 96.8526 %
Incorrectly Classified Instances 22 3.1474 %
Kappa statistic 0.9313
Mean absolute error 0.0349
Root mean squared error 0.1744
Relative absolute error 7.7298 %
Root relative squared error 36.7027 %
Total Number of Instances 699
=== Detailed Accuracy By Class ===
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.961 0.017 0.991 0.961 0.976 0.932 0.989 0.995 benign
0.983 0.039 0.929 0.983 0.956 0.932 0.989 0.976 Malignant
Weighted Avg. 0.969 0.024 0.970 0.969 0.969 0.932 0.989 0.988
=== Confusion Matrix ===
a b <-- classified as
440 18 | a = benign
4 237 | b = Malignant- Explain the meaning of the numbers in the confusion matrix.
According to the confusion matrix, there are four number, which are 440, 18, 4, 237. 440 means that there are 440 real benign cases are classified as benign class correctly. 18 means there are 18 benign cases are classified into malignant class incorrectly. 237 means that there are 237 real malignant cases are classified as malignant class correctly. 4 means there are 4 malignant cases are classified into benign class incorrectly.
Part F: Explaination (3 Marks)
Requirement
Use one record to explain how the model makes classification using the discretised dataset.
There we select the same record with the numerical part as the data for explanation.
The discreted dataset has been split into 3 bins using equal frequency approach. Here we rename the 3 bins as x,y,z, then a table will get like below.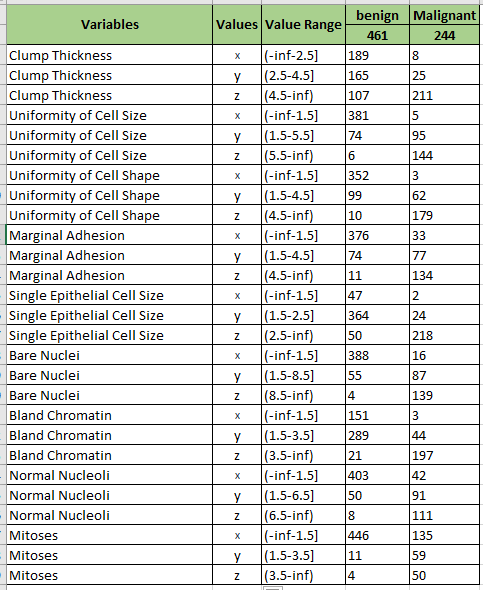
According to Image above, we could know the value of each variable. For easily to use, the variables need to rename for short.
| New Name | Original Name | original Value | Value |
|---|---|---|---|
| A | Clump Thickness | 5 | z |
| B | Uniformity of Cell Size | 1 | x |
| C | Uniformity of Cell Shape | 1 | x |
| D | Marginal Adhesion | 1 | x |
| E | Single Epithelial Cell Size | 2 | y |
| F | Bare Nuclei | 1 | x |
| G | Bland Chromatin | 3 | y |
| H | Normal Nucleoli | 1 | x |
| I | Mitoses | 1 | x |
Now, We assign
Because of
According to the data above we could get the probability for each variable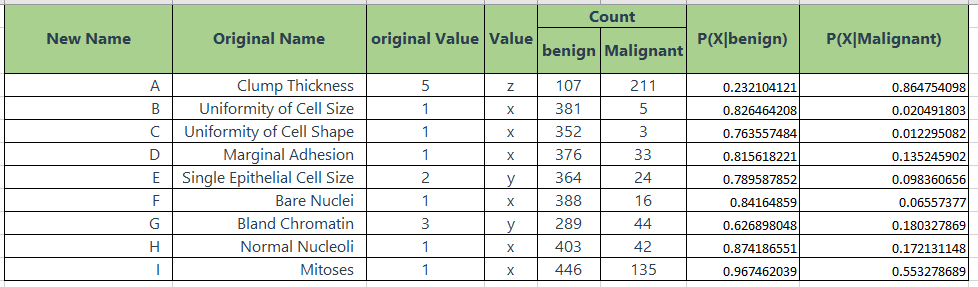
P(Class=benign) = 0.65
P(Class=malignant) = 0.35
Finally, we could calculate that
It's obviously that
So, the final result for this record is benign, it has been classifed correctlly.
References
Apache Flink® — Stateful Computations over Data Streams n.d., viewed 20 August 2023, https://flink.apache.org/.
Apache Hadoop 3.3.6 – Apache Hadoop YARN n.d., viewed 20 August 2023, https://hadoop.apache.org/docs/stable/hadoop-yarn/hadoop-yarn-site/YARN.html.
Apache HBase – Apache HBase™ Home n.d., viewed 20 August 2023, https://hbase.apache.org/.
Apache Hive n.d., viewed 20 August 2023, https://hive.apache.org/.
Apache Kylin | Analytical Data Warehouse for Big Data n.d., viewed 20 August 2023, https://kylin.apache.org/.
Apache Spark™ - Unified Engine for large-scale data analytics n.d., viewed 20 August 2023, https://spark.apache.org/.
HDFS Architecture Guide n.d., viewed 20 August 2023, https://hadoop.apache.org/docs/r1.2.1/hdfs_design.html.
Kubernetes vs YARN for scheduling Apache Spark n.d., Spot.io, viewed 20 August 2023, https://spot.io/blog/kubernetes-vs-yarn-for-scheduling-apache-spark/.
miao君 2022, 美团的大数据平台架构实践, 知乎专栏, viewed 20 August 2023, https://zhuanlan.zhihu.com/p/26359613.
Presto: Free, Open-Source SQL Query Engine for any Data n.d., viewed 20 August 2023, http://prestodb.github.io/.
Production-Grade Container Orchestration n.d., Kubernetes, viewed 20 August 2023, https://kubernetes.io/.
Sensu | How Kubernetes works n.d., Sensu, viewed 20 August 2023, https://sensu.io.
Using in MapRed n.d., viewed 20 August 2023, https://orc.apache.org/docs/mapred.html.
